Tutorial
-
- The sequence should be constituted only with the following nucleotides: A, U, T, G, C, a, u, t, g, c.
- The length of the input sequence is limited to 130 nucleotides.
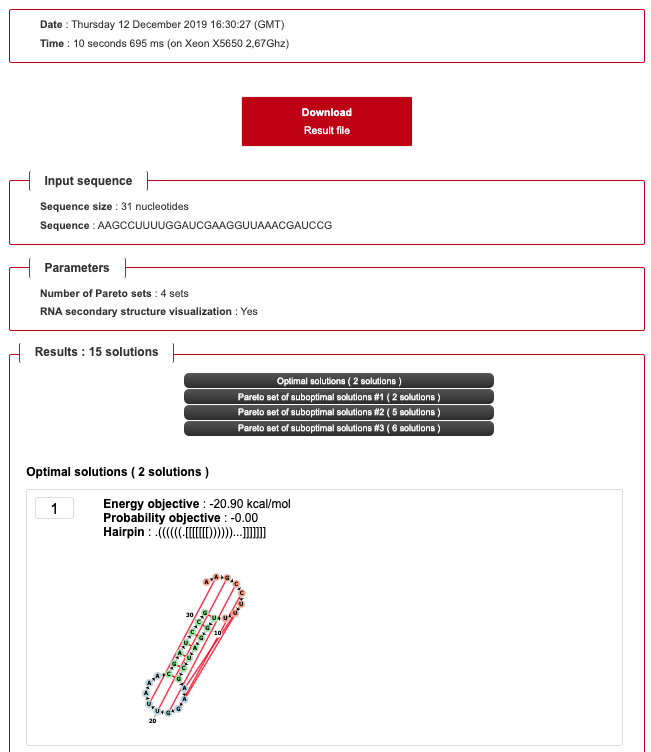
- By checking the box, you can use the following example sequence: AAGCCUUUUGGAUCGAAGGUUAAACGAUCCG
-
- Using 1 set returns the optimal solutions.
- Using more sets returns additional sets of suboptimal solutions
-
- This option enables to display RNA secondary structure diagrams :
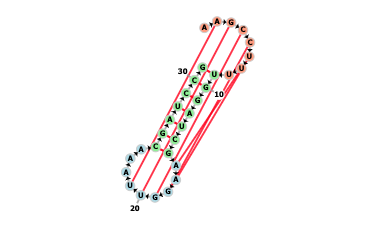
- This allows researchers to develop hypotheses about which nucleotides and base pairs are functionally relevant.
- Kerpedjiev P, Hammer S, Hofacker IL. Forna (force-directed RNA): Simple and effective online RNA secondary structure diagrams. Bioinformatics 31(20):3377-9. 2015. doi:10.1093/bioinformatics/btv372
- Press CTRL+D on the waiting page, to access results later.
- Several sets of solutions are displayed depending on the chosen number of Pareto sets.
- Each hairpins are displayed with the following informations:
- Energy objective
- Probability objective
- RNA secondary structure visualization (if enabled)
- Nucleotides are colored according to the type of structure that they are in :
- Green : Stems (canonical helices)
- Red : Multiloops (junctions)
- Yellow : Interior loops
- Blue : Hairpin loops
- Orange : 5' and 3' unpaired region
- Pseudoknots are displayed as red lines between nucleotides
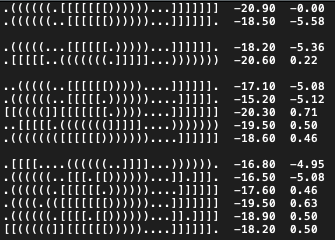
- Result file is a tab-separated text format with 3 columns:
- Hairpin
- Energy objective (kcal/mol)
- Probability objective
- Each set of solutions is separated by a empty line:
- Set #1 : Optimal solutions
- Set #2 : Pareto set of suboptimal solutions #1
- Set #3 : Pareto set of suboptimal solutions #2
- Set #4 : Pareto set of suboptimal solutions #3
- (Additional sets depending on the chosen number of Pareto sets)
| OS |
Version |
Chrome |
Firefox |
Edge |
Safari |
| Linux |
Ubuntu 18.04.3 LTS |
79.0 |
68.0 |
|
|
| macOS |
Catalina 10.15.1 |
79.0 |
72.0 |
|
no |
| Windows |
10 |
79.0 |
69.0 |
44 |
|


