RNA-SC
Tutorial
- It's the number of matchings in the reference structure, which are correctly predicted. This factor can be used for evaluating the homology degree for the two structures.
- It's the number of present matchings in the reference structure, which are not predicted.
- It's the number of present matchings in the predicted structure, which are not in the reference structure.
- As defined by Gardner and Giegerich
-
- Gardner Paul P and Giegerich Robert A comprehensive comparison of comparative RNA structure prediction approaches. BMC Bioinformatics 5:140. 2004. doi:10.1186/1471-2105-5-140
- This factor evaluate the capacity of the software to find stems.
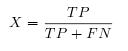
- This factor evaluate the capacity of the software to find true positive stems.
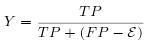
-
- This factor evaluate simultaneously the sensibility and the selectivity. The MCC requires the sequence length, in order to calculate True Negative (TN).
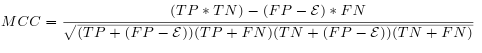
- TN corresponds to all possible matchings:
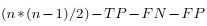
- This factor evaluate simultaneously the sensibility and the selectivity. The MCC requires the sequence length, in order to calculate True Negative (TN).
-
- Position paired format
- Bracket format
- Positions paired with the name, the sequence and the structure of the RNA.
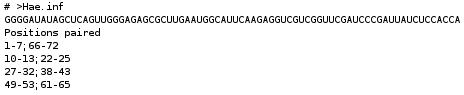
- OR
- Positions paired with only the RNA structure.

- Bracket format with the name, the sequence and the structure or the RNA.
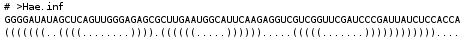
For any questions, comments or suggestions about miRNAFold, please feel free to contact: fariza.tahi@ibisc.univ-evry.fr