IRSOM2
Prediction of bifunctional RNAs
Contact:fariza.tahi@ibisc.univ-evry.fr
This website is free and open to all users and there is no login requirement.
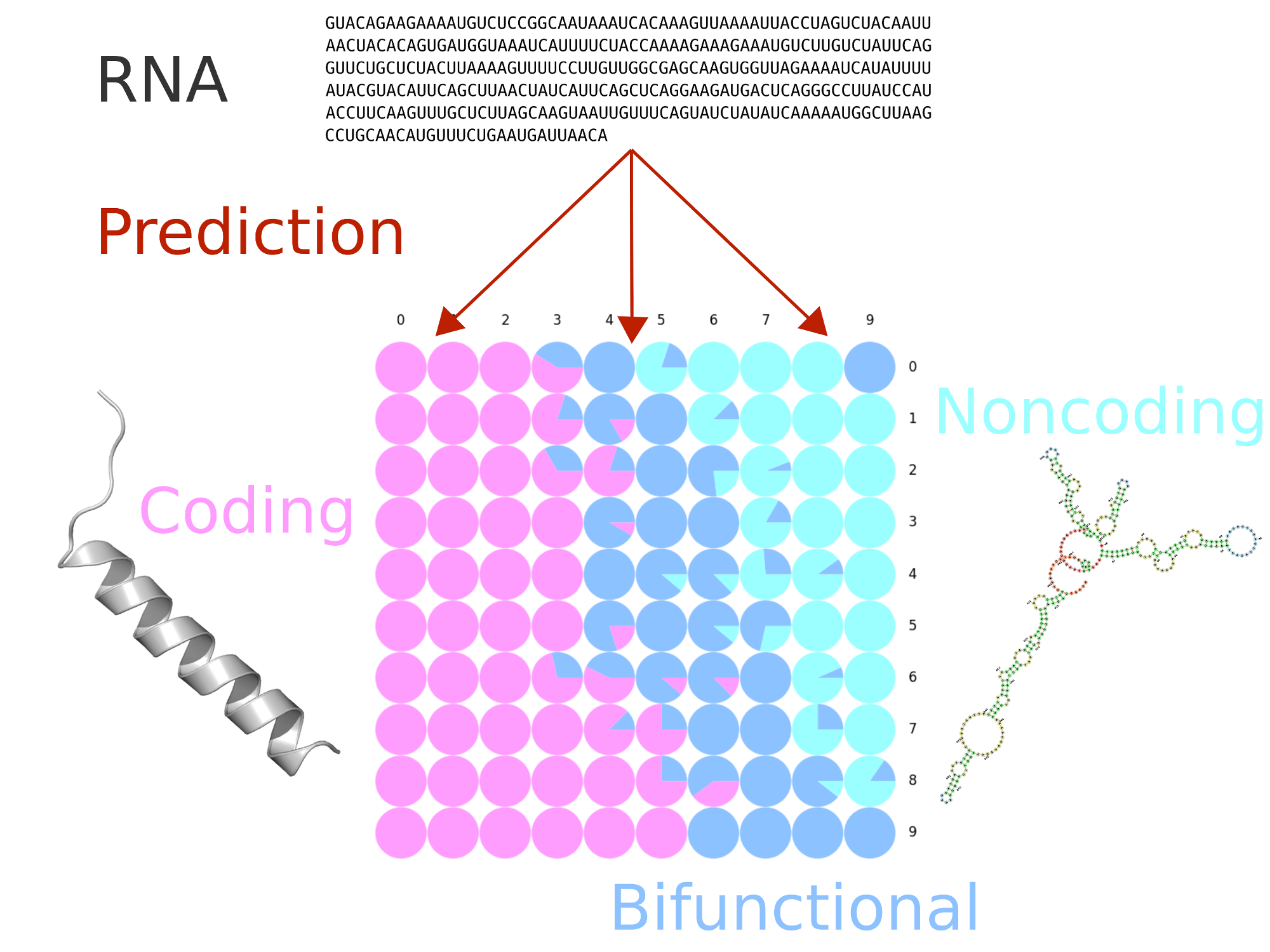
The IRSOM2 web server classifies each submitted RNA sequence into one of three categories: coding RNA, non-coding RNA, and “bifunctional RNA”. This is achieved by using self-organizing maps (SOM), taking into account features that are related to the sequence statistics and the putative open reading frames.
Pre-computed results from a dataset of 38,000 human RNA sequences: Click here
Video tutorials
Regular use
Advanced use
Model re-training
The code repository of IRSOM and the datasets used in the Bioinformatics article are available following the links below:
| OS | Version | Chrome | Firefox | Microsoft Edge | Safari |
| Linux | Ubuntu 20.04.5 LTS | 110.0 | 110.0 | 110.0 | N/A |
| MacOS | Ventura 13.2.1 | 110.0 | 110.0 | 110.0 | 16.3 |
| Windows | 10 | 110.0 | 110.0 | 110.0 | N/A |
How to cite IRSOM2 web server:
- Postic G., Tav C., Platon L., Zehraoui F., Tahi F. IRSOM2: a web server for predicting bifunctional RNAs. Nucleic Acids Research. 2023. doi:10.1093/nar/gkad381
How to cite IRSOM method:
- Platon L., Zehraoui F., Bendahmane A., Tahi F. IRSOM, a reliable identifier of ncRNAs based on supervised self-organizing maps with rejection. Bioinformatics, Volume 34, Issue 17, 01 September 2018, Pages i620–i628. doi:10.1093/bioinformatics/bty572
For any questions, comments or suggestions about IRSOM, please feel free to contact: fariza.tahi@ibisc.univ-evry.fr