RNANet
RNANet
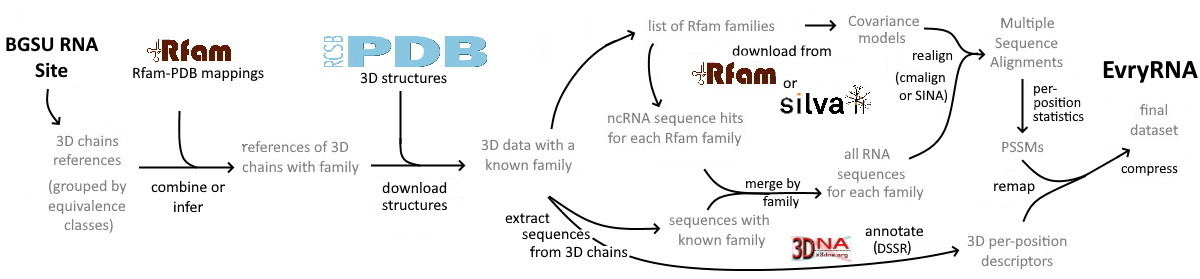
A major part of any data-science work consists in finding appropriate data which contains enough signal to tackle the problem we are interested in. Then, cleaning the data to ensure uniformity of the measures, compatibility of the various data sources and protocols, and a reasonable amount of noise is sometimes the most time-consuming step.
Here we propose a first attempt of standardized and automatically generated dataset dedicated to non-coding RNA combining together:
- Sequence, including modified bases,
- Secondary structure, including non-canonical basepairs and multipairs,
- Standardized 3D structures and 3D geometrical descriptors and annotations,
- Homology information like nucleotide frequencies and covarianc models for every position in a 3D chain, sequence consensus, and sequence alignments.
We hope this dataset will speed-up advances in machine-learning based approaches for RNA secondary and/or 3D structure prediction, by avoiding spending time on data gathering and cleaning.
Downloads
SQLite3 Database
Text files (CSV)
Git repository
- RNANet.db : A SQLite3 database containing all the information. You might want to query it to build your own sub-datasets.
- Text-files : CSV files summarizing the information for every RNA 3D chain (1 file per 3D chain mapped on a Rfam family)
Extract the archives using commands gunzip RNANet.db.gz to recreate RNANet.db and tar -xvzf RNANET_datapoints_latest.tar.gz to recreate a folder of text files.
Additional files and meta-data:
These might be useful to you to assert the dataset quality, or perform further filtering.
- Sequence alignments by family (tar.gz archive) : FASTA files containing aligned sequences of the portions of RNA chains which have a 3D structure and are mapped to an Rfam family.
- Normalized 3D mmCIF files containing non-coding RNA (tar.gz archive):
One chain per .cif file, renumbered, and coherent with the database.
Download all RNA, or fragments mapped to Rfam families. - summary.csv : Additional information about the previous RNA chains (date of publication, resolution, basepair types counts)
- families.csv : Additional information about the Rfam RNA families used (number of 3D chains, number of homologous sequences)
- frequencies.csv : Nucleotide frequencies by RNA family, including modified bases
- pair_types.csv : Basepair-type frequencies by RNA family, including only intra-chain base-base interactions, in Leontis-Westhof nomenclature
You can also browse all past releases of the flat-text files (approx. one per month)
Updates
RNANet is updated monthly to take newer structures from the BGSU representative sets into account.
RNANet is updated from scratch twice a year to take into account new sequences from Rfam, and updates in covariance models or PDB structures.
Documentation
You can read our OpenAccess paper. Extensive documentation can also be found in the code repository:
- Installation and run guide
- Database documentation
- FAQ
- Known issues and known feature requests
- Warning and errors help list
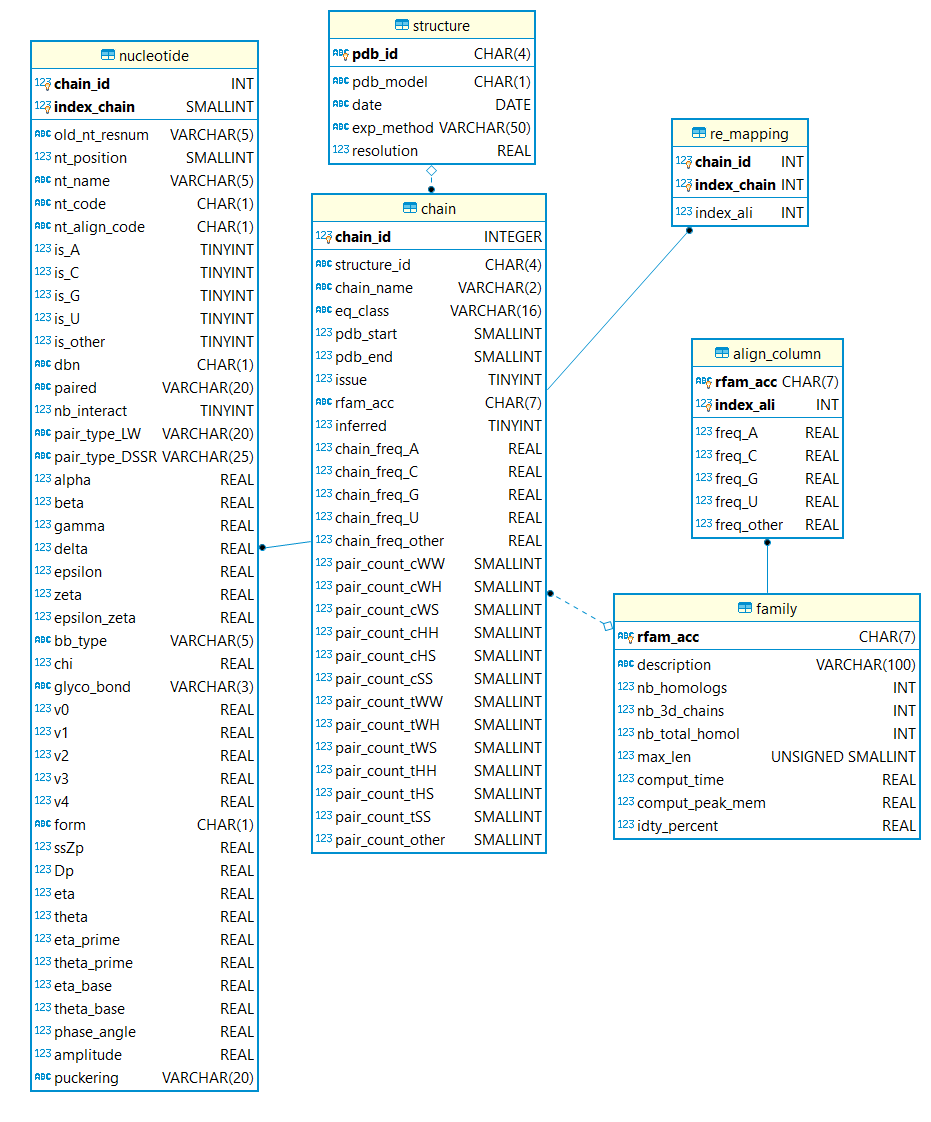
For each RNA chain available in 3D and mapped to a RNA family, we provide the following list of descriptors:
Descriptor |
Label |
Type |
| Index of the residue in the chain (from 1 to N) | index_chain | int >= 1 |
| Position of the nucleotide in the Rfam family model (covariance model). NaN in insertions. | cm_coord | int >= 1 |
| Position of the nucleotide in the multiple-sequence alignment of the 3D chains mapped to this family. | index_small_ali | int >= 1 |
| Index of the residue in the source mmCIF file | old_nt_resnum | int >= 1 |
| Position of the nucleotide in the chain, normalized by its length (value between 0 and 1) | nt_position | float |
| Nucleotide name, including modified bases (like 5MC) | nt_name | str |
| One-letter name. Lowercase "acgtu" letters are used for modified "ACGTU" bases | nt_code | char |
| Letter used for sequence alignment (A,C,G,U,N, or -) | nt_align_code | char |
| One-hot encoded sequence. 'other' contains gaps, unknown and modified nucleotides | is_A, is_C, is_G, is_U, is_other | 0 or 1 |
| Nucleotide frequencies (PSSM) at the current position in this RNA family | freq_A, freq_C, freq_G, freq_U, freq_other |
float |
| Frequency of gaps at this position in the alignment (between (0.0 and 1.0). | gap_percent | float |
| Consensus nucleotide from the alignment at this position (A,C,G,U,N or -). | consensus | char |
| Secondary structure in dot-bracket notation of this position | dbn | char |
| Zero, or comma-separated values of index_chain of the nucleotide(s) which is(are) paired with this one. Canonical (Watson-Crick or Wobble) basepairs are first in the list. | paired | int, int, ... |
| Number of bases interacting with this one | nb_interact | int >= 0 |
| Type of basepair in Leontis-Westhof nomenclature (comma-separated list) | pair_type_LW | str, str, ... |
| Type of basepair in DSSR nomenclature (comma-separated list) | pair_type_DSSR | str, str, ... |
| The six torsion angles of the backbone, from 5' to 3', between 0 and 2pi | alpha, beta, gamma, delta, epsilon, zeta | float (rad) |
| Difference between epsilon and zeta torsion angles | epsilon_zeta | float (rad) |
| Conformation of the backbone | bb_type | BI, BII, '..', or 'n/a' |
| Chi torsion angle (between ribose and base) | chi | float (rad) |
| Conformation of the sugar with respect to the base (depends on Chi) | glyco_bond | syn or anti |
| Torsion angles of the ribose cycle | v0, v1, v2, v3, v4 | float (rad) |
| If the nucleotide is involved in a stem, the stem type | form | A, B, Z or '.' |
| Z-coordinate of the 3' phosphorus atom with reference to the 5' base plane | ssZp | float |
| Perpendicular distance of the 3' P atom to the glycosidic bond | Dp | float |
| Pseudotorsions between P and C1' | eta, theta | float (rad) |
| Pseudotorsions between P and C4' | eta_prime, theta_prime | float (rad) |
| Pseudotorsions between P and the base center | eta_base, theta_base | float (rad) |
| Conformation of the ribose cycle | phase_angle | float (rad) |
| Amplitude of the sugar puckering | amplitude | float |
| Conformation of the ribose cycle (10 classes corresponding to specific ranges of phase) | puckering | str |
More descriptors are available in the SQL tables. The database scheme is illustrated below:
Dataset filtering quick example
It is possible to build sub-datasets by querying the results/RNANet.db file. We provide examples using Python3 and the sqlite3 package:
import sqlite3
import pandas as pd
with sqlite3.connect("results/RNANet.db) as connection:
df = pd.read_sql("""SELECT structure_id, chain_name
FROM chain JOIN structure ON chain.structure_id = structure.pdb_id
WHERE resolution < 4.0 ORDER BY date ASC;""", con=connection)
df.to_csv("my_custom_results.csv")
More examples of SQL queries can be found in the README.md file on the IBISC forge, see section How to further filter the dataset.
References
How to cite RNANet:
- Louis Becquey, Eric Angel, and Fariza Tahi, RNANet: an automatically built dual-source dataset integrating homologous sequences and RNA structures, Bioinformatics, 2020, btaa944, DOI
Additional references:
- The "ProteinNet" philosophy which inspired this work:
- AlQuraishi, M. (2019b). ProteinNet: A standardized data set for machine learning of protein structure. BMC Bioinformatics, 20(1), 311
- If you use our annotations by DSSR, you might want to cite:
- Lu, X.-J.et al.(2015). DSSR: An integrated software tool for dissecting the spatial structure of RNA. Nucleic Acids Research, 43(21), e142–e142.
- If you use our multiple sequence alignments and homology data, you might want to cite:
- Pruesse, E.et al.(2012). Sina: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics, 28(14), 1823–1829
- Nawrocki, E. P. and Eddy, S. R. (2013). Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics, 29(22), 2933–2935.