Tutorial
-
- The sequence should be constituted only with the following nucleotides: A, T, C, G, N, a, t, c, g, n.
- Single sequence mode: One sequence in FASTA file format, the first line of your file must start with ">"
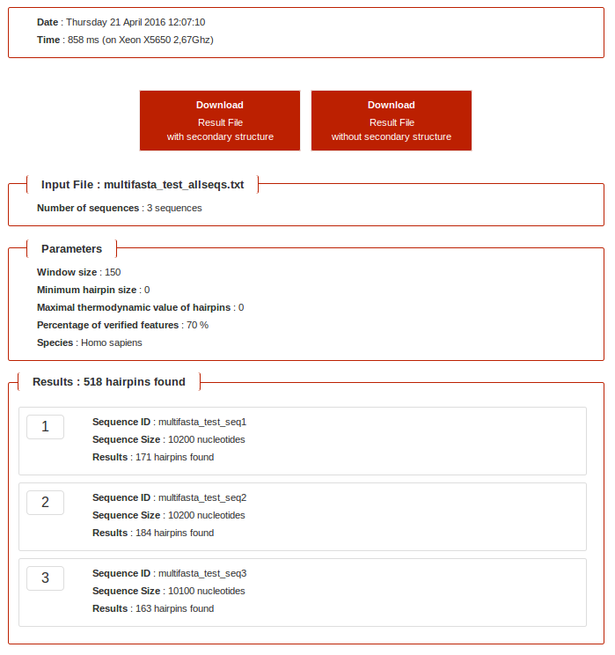
- Multifasta mode: Input file could be constituted by several sequences in FASTA format
-
- The maximal size of the pre-miRNA that miRNAFold can find.
- This size is limited to 1000 nucleotides.
-
- The minimal size of the pre-miRNA secondary structure.
-
- The maximal thermodynamic value ?G of the miRNAFold predicted hairpin.
-
- miRNAFold calculates and compares lots of values to the specie features (hairpin size, G+C percent, ...).
- Each feature has a minimal and a maximal threshold.
- "Percentage of verified features" is the percentage of calculated value between the two thresholds of a specie feature.
- miRNAFold can be run on all genomes.
- Available 78 species are:
- Homo sapiens
- Acyrthosiphon pisum
- Aedes aegypti
- Amborella trichopoda
- Anolis carolinensis
- Apis mellifera
- Arabidopsis lyrata
- Arabidopsis thaliana
- Bombyx mori
- Bos taurus
- Brachypodium distachyon
- Branchiostoma belcheri
- Branchiostoma floridae
- Brugia malayi
- Caenorhabditis brenneri
- Caenorhabditis briggsae
- Caenorhabditis elegans
- Caenorhabditis remanei
- Canis familiaris
- Capitella teleta
- Capra hircus
- Ciona intestinalis
- Cricetulus griseus
- Cucumis melo
- Cyprinus carpio
- Danio rerio
- Drosophila melanogaster
- Drosophila pseudoobscura
- Drosophila simulans
- Drosophila virilis
- Eptesicus fuscus
- Equus caballus
- Fugu rubripes
- Gallus gallus
- Glycine max
- Gorilla gorilla
- Gossypium raimondii
- Haemonchus contortus
- Ictalurus punctatus
- Linum usitatissimum
- Macaca mulatta
- Malus domestica
- Manihot esculenta
- Medicago truncatula
- Monodelphis domestica
- Mus musculus
- Nematostella vectensis
- Nicotiana tabacum
- Ophiophagus hannah
- Ornithorhynchus anatinus
- Oryza sativa
- Oryzias latipes
- Ovis aries
- Pan troglodytes
- Panagrellus redivivus
- Petromyzon marinus
- Physcomitrella patens
- Plutella xylostella
- Pongo pygmaeus
- Populus trichocarpa
- Pristionchus pacificus
- Prunus persica
- Rattus norvegicus
- Salmo salar
- Schistosoma mansoni
- Schmidtea mediterranea
- Solanum tuberosum
- Sorghum bicolor
- Strongyloides ratti
- Sus scrofa
- Taeniopygia guttata
- Tetraodon nigroviridis
- Tribolium castaneum
- Triticum aestivum
- Tupaia chinensis
- Vitis vinifera
- Xenopus tropicalis
- Zea mays
- Press CTRL + D on the waiting page, to access results later.
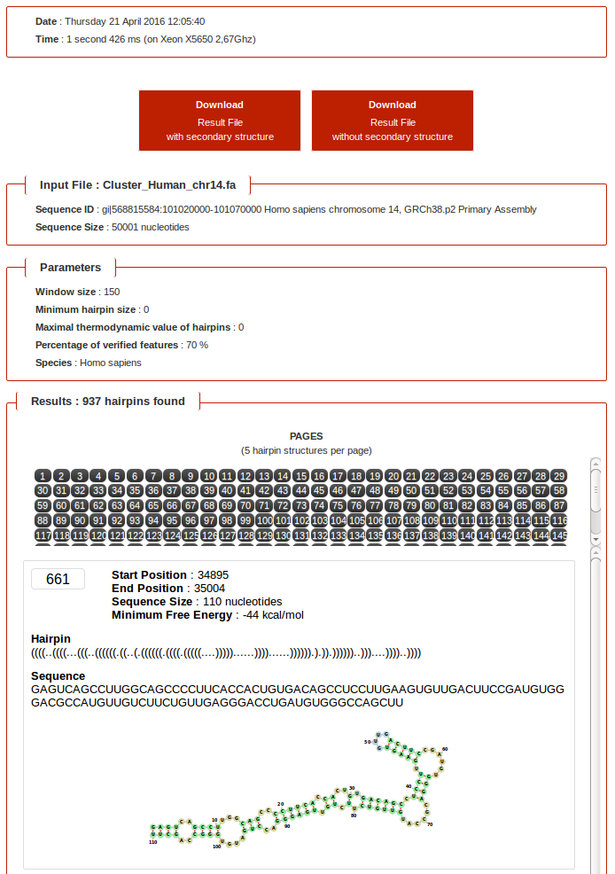
- Each hairpins are displayed with the following informations:
- Start Position
- End Position
- Sequence Size
- Minimum Free Energy
- Hairpin
- Sequence
- Secondary Structure Visualization
- NB: In multifasta mode, only size and number of hairpins found for each sequence are displayed.
- This allows researchers to develop hypotheses about which nucleotides and base pairs are functionally relevant.
- Kerpedjiev P, Hammer S, Hofacker IL. Forna (force-directed RNA): Simple and effective online RNA secondary structure diagrams. Bioinformatics 31(20):3377-9. 2015. doi:10.1093/bioinformatics/btv372
- NB: This option isn't available for multifasta mode
- Two types of result file are available: with or without secondary structure